
Principal Investigator
To maintain cell and tissue function and homeostasis in multicellular organisms, cells need to constantly respond to different signals and adapt their behaviors. This is achieved through precise regulation of signaling pathways that govern gene expression.
The Uhlirova Lab explores the pleiotropic functions of stress-inducible signaling pathways and the downstream mechanisms orchestrating gene expression. We investigate how diverse stress signals modulate transcription factor and spliceosome activities during development and physiology. Additionally, we examine how dysfunction in transcription and splicing machinery impacts cell and tissue behaviors, as well as organismal homeostasis during aging and disease. Our approach integrates functional genetics in Drosophila and mouse models with a diverse array of techniques, including molecular and cell biology, biochemistry, multi-omic approaches, and advanced microscopy.
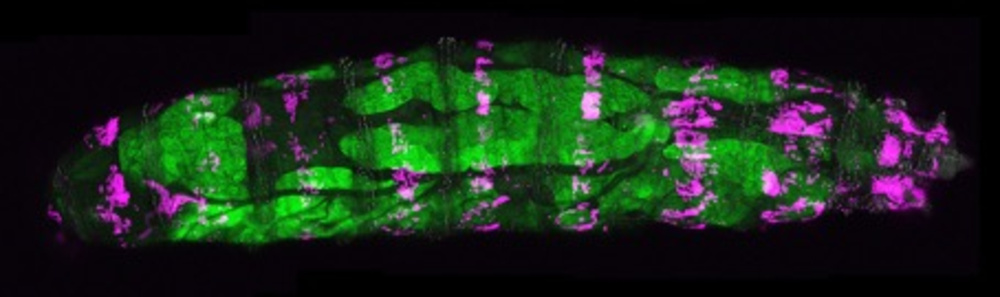
© Gábor Csordás
The ability of cells to sense and interpret various stress signals, generating meaningful biological responses by coordinating transcription and pre-mRNA splicing is critical for the development, maintenance, and healthspan of multicellular organisms.
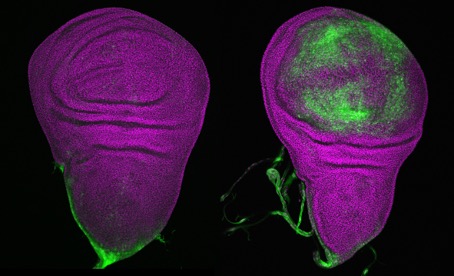
© Mirka Uhlirova
Transcription and pre-mRNA splicing are essential steps in the regulation of gene expression in multicellular organisms. Their close interplay enables the generation of specific gene expression patterns required for the differentiation of functionally diverse cell types, but also allows the fine-tuning of their responses to various cues, providing a dynamic and adaptive ability to navigate their ever-changing environment.
Our goal is to understand how co-transcriptional splicing governs essential cellular behaviors, such as growth, proliferation, cell death, and cell migration both in physiological conditions and under stress. We particularly focus on epithelia and immune cells and their interactions. We investigate the link between pre-mRNA splicing and the maintenance of transcriptome and genome integrity.
In summary, our research aims to unravel the intricate connections between transcription, pre-mRNA splicing, and their collective impact on cellular behavior, tissue homeostasis, aging, and disease development. By elucidating these processes, we seek to provide valuable insights into the function of fundamental biological machines, with potential implications for therapeutic intervention.
We demonstrated the pivotal role of spliceosome biogenesis in the development, tissue maintenance, and overall healthspan of multicellular organisms (Erkelenz et al., 2021, NAR) and identified C/EBP transcription factors as key drivers of a stress response program and senescent-like state triggered by spliceosome dysfunction (Stankovic et al., 2024, NAR).
We established the Drosophila model to study the mechanisms underlying an autosomal dominant form of an eye-blinding degenerative disease retinitis pigmentosa caused by mutations in a core splicing factor Prp8 (Stankovic et al., 2020, DMM).
We defined a physiological role for the transcription factor Ets21c, the fly ortholog of mammalian proto-oncogenes ERG and FLI-1, in the regulation of the regenerative program in the Drosophila adult intestine and highlighted its role in the trade-off mechanisms between stress resilience and longevity (Mundorf et al., 2019, Cell Rep.).
Our research has provided novel insights into how tumor cells exploit mechanosensitive cellular machinery to promote malignancy (Külshammer et al., 2022, FEBS J).
Finally, our interest in the mechanisms of inter-organ communication identified the interaction between a phagocytic receptor Eater and an extracellular matrix component Multiplexin, the fly ortholog of human Collagen XV/XVIII, to be necessary and sufficient for blood cell homing and their interactions with hematopoietic microenvironments (Csordas et al., 2020, Elife).

Principal Investigator